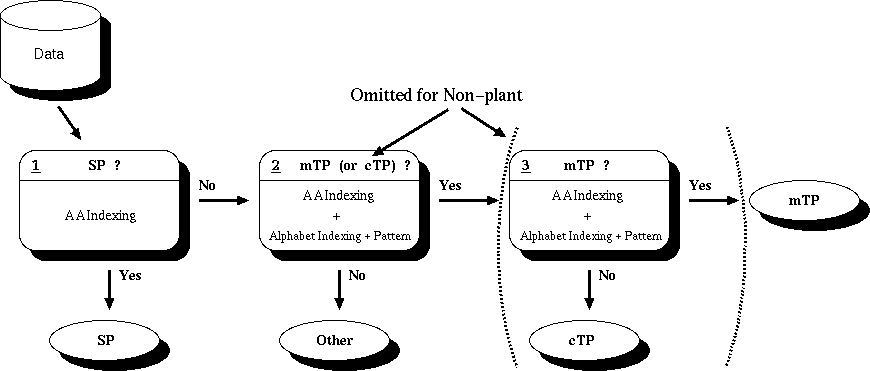
iPSORT is a subcellular localization site predictor for N-terminal sorting signals. Given a protein sequence , it will predict whether it contains a Signal Peptide (SP), Mitochondrial Targeting Peptide (mTP), or Chloroplast Transit Peptide (cTP).
The structure of iPSORT is simply a decision list consisting of 3 nodes. (2 for non-plant)
(click on image to enlarge)
At the 1st node, the protein sequence is checked if it is a Signal Peptide (SP) or not. If it is predicted as SP, then the output is simply "SP".
At the second node, the protein sequence is judged if it is either a mitochondrial targeting peptide (mTP), or chloroplast transit peptide (cTP). If it is determined not to be either of them, the sequence is predicted to be "Other". (For non-plant sequences, this will be the final node).
At the last node, the protein sequence is judged if it is a mitochondrial targeting peptide or not. If yes, then "mTP" is the output, and if no, "cTP".
The rules deciding whether or not the given signals contain a certain signal consists of two elements: an amino acid index rule, and an alphabet indexing+pattern rule. (except for SP with only an amino acid index rule). To be judged "yes" at each node, the input amino acid sequence must satisfy both of the two rules (except SP). They are explained below.
An amino acid index is a mapping from an amino acid to a numerical value. For a given amino acid, its amino acid index represents some biochemical property of the amino acid.
Using these amino acid index values, the average amino acid index value of certain substrings of the input amino acid sequence are calculated at each node. To be judged "yes" at a given node, the average must exceed (or be less than) a certain threshold. The indices, substrings (shown as intervals), and thresholds, which are used for iPSORT, are given in the next table.
| Data Type | Node | Amino Acid Index | Substring Interval | Threshold |
|---|---|---|---|---|
| Plant | 1 | Hydropathy Index | [6, 25] | avg >= 0.9225 |
| 2 | Negative Charge | [1, 30] | avg < 0.083 | |
| 3 | Isoelectric Point | [1, 15] | avg >= 0.621 | |
| Non-plant | 1 | Hydropathy Index | [6, 20] | avg >= 0.953 |
| 2 | Net Charge | [1, 30] | avg >= 0.083 |
The values for each amino acid for the indices are given in the table below.
| Amino Acid Index | Amino Acid | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | R | N | D | C | Q | E | G | H | I | L | K | M | F | P | S | T | W | Y | V | |
| Hydropathy Index | 1.8 | -4.5 | -3.5 | -3.5 | 2.5 | -3.5 | -3.5 | -0.4 | -3.2 | 4.5 | 3.8 | -3.9 | 1.9 | 2.8 | -1.6 | -0.8 | -0.7 | -0.9 | -1.3 | 4.2 |
| Negative Charge | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Isoelectric Point | 6.00 | 10.76 | 5.41 | 2.77 | 5.05 | 5.65 | 3.22 | 5.97 | 7.59 | 6.02 | 5.98 | 9.74 | 5.74 | 5.48 | 6.30 | 5.68 | 5.66 | 5.89 | 5.66 | 5.96 |
| Net Charge | 0.0 | 1.0 | 0.0 | -1.0 | 0.0 | 0.0 | -1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
An alphabet indexing can be considered as a discrete, non-ordered version of an amino acid index. Each amino acid is converted to a fewer class of characters (to characters '0', '1', '2' in our case). The original amino acid sequence is converted to a string of 0s, 1s, and 2s. To be judged "yes" at a given node, the converted sequence must match a certain pattern, within a certain substring. We also allow mismatches in the matching.
The parameters used in iPSORT are summarized below.
| Data Type | Node | Alphabet Indexing | Substring | Pattern | Mismatch | ||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | |||||
| Plant | 1 | --- | --- | --- | --- | --- | --- |
| 2 | DEGHKN | IR | ACFLMPQSTVWY | [1,30] | 22121222 | up to 2 insertion/deletion | |
| 3 | ACDEFGHLMNQSTVWY | KR | IP | [1,15] | 100100110 | up to 3 insertion/deletion | |
| Non-plant | 1 | --- | --- | --- | --- | --- | --- |
| 2 | DEGHKN | IR | ACFLMPQSTVWY | [1,30] | 221121122 | up to 3 insertion/deletion | 3 | --- | --- | --- | --- | --- | --- |
Using the plant data set used to train TargetP, we searched for the best combination of: which substring interval to look at, what amino acid indices to use, what thresholds, what alphabet indexing, and what patterns to use.
For each pair of sorting signal types (including Other), all substrings of [5n+1, 5k] (where n, k are integers 0 <= n <= 8, 1 <= k <= 8), all 434 amino acid indices in the AAIndex Database (+ 20 'counting' indices where one amino acid is given value '1' and the rest '0'), all possible thresholds, all patterns of length 3-12 with mismatches 0-3 were searched. For alphabet indexing, an exhaustive search was not feasible, and a local search was conducted a many number of times.
The construction of the decision list was done in a greedily, 'filtering' well classified signals first.
The scores for a 5-fold cross validation are shown in the following table, in comparison with TargetP. The score used is Matthews correlation coefficient (MCC) given by (tp*tn - fp*fn)/sqrt((tp+fn)(tp+fp)(tn+fp)(tn+fn)) where tp, fp, tn, fn are the # of true positives, false positives, true negatives, and false negatives, respectively.For the non-plant dataset, the same decision list structure and interval for alphabet indexing was used. The intervals and thresholds for amino acid indexing and patterns were trained.
| Data Set | True Category | # of sequences | Predicted Category | Sensitivity | MCC | |||
|---|---|---|---|---|---|---|---|---|
| cTP | mTP | SP | Other | |||||
| Plant | cTP | 141 | 112 (120) | 15 (14) | 0 (2) | 14 (5) | 0.79 (0.85) | 0.64 (0.72) |
| mTP | 368 | 41 (41) | 304 (300) | 9 (9) | 14 (18) | 0.83 (0.82) | 0.79 (0.77) | |
| SP | 269 | 16 (2) | 8 (7) | 237 (245) | 8 (15) | 0.88 (0.91) | 0.89 (0.90) | |
| Other | 162 | 13 (10) | 6 (13) | 2 (2) | 141 (137) | 0.87 (0.85) | 0.80 (0.77) | |
| Specificity | 0.62 (0.69) | 0.91 (0.90) | 0.96 (0.96) | 0.80 (0.78) | -- | -- | ||
| Non-plant | mTP | 371 | -- | 293 (330) | 14 (9) | 64 (32) | 0.79 (0.82) | 0.70 (0.73) |
| SP | 715 | -- | 19 (13) | 620 (683) | 76 (19) | 0.87 (0.91) | 0.85 (0.92) | |
| Other | 1652 | -- | 108 (152) | 48 (49) | 1496 (1451) | 0.91 (0.85) | 0.77 (0.82) | |
| Specificity | -- | 0.70 (0.67) | 0.91 (0.92) | 0.91 (0.97) | -- | -- | ||